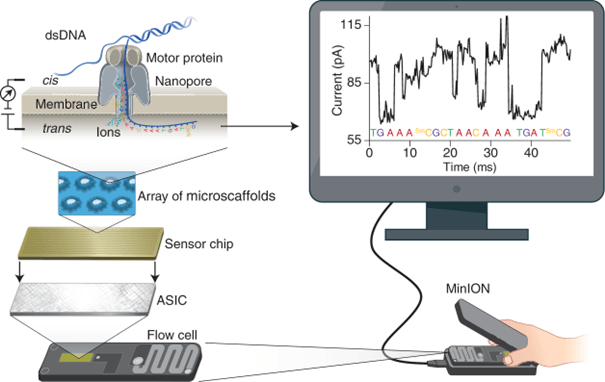
MinION: the new portable mini-sequencer at GEVES
At GEVES in Beaucouzé, the MinION Mk1B high throughput sequencer has been installed. This 3rd generation sequencer, which fits in one hand, can be used for real-time sequencing of long DNA or RNA fragments.
Following this acquisition and with the precious advice of Jessica Dittmer (INRAE – IRHS), the GEVES successfully sequenced its first bacterial strain: the Xanthomonas campestris pv. raphani 756C reference strain. The newly sequenced genome shows 99.91% identity with the genome available in public databases. As a result, GEVES will be able to characterise the collection of pathogens it holds in the GEVES pathology laboratory.
In addition, several development areas are envisaged to integrate the MinION into the missions of the GEVES Detection Unit, such as the identification of genetic variation, sample multiplexing, GMO detection or the detection of multiple pathogens in a seed sample in a single analysis using metabarcoding.
For this last area of development, a CASDAR project coordinated by GEVES, in partnership with INRAE, SFR QuaSaV-ANAN and seed companies (HM.Clause, Vilmorin-Mikado and Sakata Europe), plans to test this detection method: the SeqDetectVeg project.